Complex Numbers, Mean, and Classification#
Complex numbers
Absolute value
Evenly spaced numbers
Mean
Arithmetic mean
Geometric mean
Harmonic mean
Classification
Complex Numbers#
A complex number \(z\) is represented by \(z = x + j* y\), where \(x\) and \(y\) are real numbers.
c1 = complex(0, 0)
print(c1)
0j
x = 2
y = 3
c2 = complex(x, y)
print(c2)
(2+3j)
Absolute value#
Absolute value of a complex number \(z = x + j* y\) is obtained by the formula \(\sqrt {(x^2 + y^2)}\)
from math import sqrt
absc2 = sqrt(x**2 + y**2)
print(absc2)
3.605551275463989
Another way is to use the function \(abs()\).
absc2 = abs(c2)
print(absc2)
3.605551275463989
absc1 = abs(c1)
print(absc1)
0.0
Evenly spaced numbers#
import numpy as np
np.linspace(1.0, 5.0, num=5)
array([1., 2., 3., 4., 5.])
import numpy as np
np.linspace(1.0, 5.0, num=10)
array([1. , 1.44444444, 1.88888889, 2.33333333, 2.77777778,
3.22222222, 3.66666667, 4.11111111, 4.55555556, 5. ])
Mean#
Arithmetic mean#
If \(a_1, a_2, \ldots, a_n\) are numbers, then the arithmetic mean is obtained by:
\[A=\frac{1}{n}\sum_{i=1}^n a_i=\frac{a_1+a_2+\cdots+a_n}{n}\]
num = np.linspace(1.0, 100.0, num=100)
a = np.mean(num)
print(a)
50.5
Geometric Mean#
If \(a_1, a_2, \ldots, a_n\) are numbers, then the arithmetic mean is obtained by:
\[\left(\prod_{i=1}^n a_i\right)^\frac{1}{n} = \sqrt[n]{a_1 a_2 \cdots a_n}\]
from scipy.stats import gmean
g = gmean(num)
print(g)
37.992689344834304
Harmonic Mean#
If \(a_1, a_2, \ldots, a_n\) are numbers, then the harmonic mean is obtained by:
\[H = \frac{n}{\frac1{a_1} + \frac1{a_2} + \cdots + \frac1{a_n}} = \frac{n}{\sum\limits_{i=1}^n \frac1{a_i}} = \left(\frac{\sum\limits_{i=1}^n a_i^{-1}}{n}\right)^{-1}.\]
from scipy.stats import hmean
h = hmean(num)
print(h)
19.277563597396004
Classification#
Understanding IRIS dataset#
from sklearn.datasets import load_iris
data = load_iris()
# Target Classes and Names
for target, name in enumerate(data.target_names):
print(target, name)
0 setosa
1 versicolor
2 virginica
# Features
for feature in data.feature_names:
print(feature)
sepal length (cm)
sepal width (cm)
petal length (cm)
petal width (cm)
X = data.data
Y = data.target
print(f"Data size: {len(X)}")
Data size: 150
# Printing the data
for x, y in zip(X, Y):
print(f" {x}: {y} ({data.target_names[y]})")
[5.1 3.5 1.4 0.2]: 0 (setosa)
[4.9 3. 1.4 0.2]: 0 (setosa)
[4.7 3.2 1.3 0.2]: 0 (setosa)
[4.6 3.1 1.5 0.2]: 0 (setosa)
[5. 3.6 1.4 0.2]: 0 (setosa)
[5.4 3.9 1.7 0.4]: 0 (setosa)
[4.6 3.4 1.4 0.3]: 0 (setosa)
[5. 3.4 1.5 0.2]: 0 (setosa)
[4.4 2.9 1.4 0.2]: 0 (setosa)
[4.9 3.1 1.5 0.1]: 0 (setosa)
[5.4 3.7 1.5 0.2]: 0 (setosa)
[4.8 3.4 1.6 0.2]: 0 (setosa)
[4.8 3. 1.4 0.1]: 0 (setosa)
[4.3 3. 1.1 0.1]: 0 (setosa)
[5.8 4. 1.2 0.2]: 0 (setosa)
[5.7 4.4 1.5 0.4]: 0 (setosa)
[5.4 3.9 1.3 0.4]: 0 (setosa)
[5.1 3.5 1.4 0.3]: 0 (setosa)
[5.7 3.8 1.7 0.3]: 0 (setosa)
[5.1 3.8 1.5 0.3]: 0 (setosa)
[5.4 3.4 1.7 0.2]: 0 (setosa)
[5.1 3.7 1.5 0.4]: 0 (setosa)
[4.6 3.6 1. 0.2]: 0 (setosa)
[5.1 3.3 1.7 0.5]: 0 (setosa)
[4.8 3.4 1.9 0.2]: 0 (setosa)
[5. 3. 1.6 0.2]: 0 (setosa)
[5. 3.4 1.6 0.4]: 0 (setosa)
[5.2 3.5 1.5 0.2]: 0 (setosa)
[5.2 3.4 1.4 0.2]: 0 (setosa)
[4.7 3.2 1.6 0.2]: 0 (setosa)
[4.8 3.1 1.6 0.2]: 0 (setosa)
[5.4 3.4 1.5 0.4]: 0 (setosa)
[5.2 4.1 1.5 0.1]: 0 (setosa)
[5.5 4.2 1.4 0.2]: 0 (setosa)
[4.9 3.1 1.5 0.2]: 0 (setosa)
[5. 3.2 1.2 0.2]: 0 (setosa)
[5.5 3.5 1.3 0.2]: 0 (setosa)
[4.9 3.6 1.4 0.1]: 0 (setosa)
[4.4 3. 1.3 0.2]: 0 (setosa)
[5.1 3.4 1.5 0.2]: 0 (setosa)
[5. 3.5 1.3 0.3]: 0 (setosa)
[4.5 2.3 1.3 0.3]: 0 (setosa)
[4.4 3.2 1.3 0.2]: 0 (setosa)
[5. 3.5 1.6 0.6]: 0 (setosa)
[5.1 3.8 1.9 0.4]: 0 (setosa)
[4.8 3. 1.4 0.3]: 0 (setosa)
[5.1 3.8 1.6 0.2]: 0 (setosa)
[4.6 3.2 1.4 0.2]: 0 (setosa)
[5.3 3.7 1.5 0.2]: 0 (setosa)
[5. 3.3 1.4 0.2]: 0 (setosa)
[7. 3.2 4.7 1.4]: 1 (versicolor)
[6.4 3.2 4.5 1.5]: 1 (versicolor)
[6.9 3.1 4.9 1.5]: 1 (versicolor)
[5.5 2.3 4. 1.3]: 1 (versicolor)
[6.5 2.8 4.6 1.5]: 1 (versicolor)
[5.7 2.8 4.5 1.3]: 1 (versicolor)
[6.3 3.3 4.7 1.6]: 1 (versicolor)
[4.9 2.4 3.3 1. ]: 1 (versicolor)
[6.6 2.9 4.6 1.3]: 1 (versicolor)
[5.2 2.7 3.9 1.4]: 1 (versicolor)
[5. 2. 3.5 1. ]: 1 (versicolor)
[5.9 3. 4.2 1.5]: 1 (versicolor)
[6. 2.2 4. 1. ]: 1 (versicolor)
[6.1 2.9 4.7 1.4]: 1 (versicolor)
[5.6 2.9 3.6 1.3]: 1 (versicolor)
[6.7 3.1 4.4 1.4]: 1 (versicolor)
[5.6 3. 4.5 1.5]: 1 (versicolor)
[5.8 2.7 4.1 1. ]: 1 (versicolor)
[6.2 2.2 4.5 1.5]: 1 (versicolor)
[5.6 2.5 3.9 1.1]: 1 (versicolor)
[5.9 3.2 4.8 1.8]: 1 (versicolor)
[6.1 2.8 4. 1.3]: 1 (versicolor)
[6.3 2.5 4.9 1.5]: 1 (versicolor)
[6.1 2.8 4.7 1.2]: 1 (versicolor)
[6.4 2.9 4.3 1.3]: 1 (versicolor)
[6.6 3. 4.4 1.4]: 1 (versicolor)
[6.8 2.8 4.8 1.4]: 1 (versicolor)
[6.7 3. 5. 1.7]: 1 (versicolor)
[6. 2.9 4.5 1.5]: 1 (versicolor)
[5.7 2.6 3.5 1. ]: 1 (versicolor)
[5.5 2.4 3.8 1.1]: 1 (versicolor)
[5.5 2.4 3.7 1. ]: 1 (versicolor)
[5.8 2.7 3.9 1.2]: 1 (versicolor)
[6. 2.7 5.1 1.6]: 1 (versicolor)
[5.4 3. 4.5 1.5]: 1 (versicolor)
[6. 3.4 4.5 1.6]: 1 (versicolor)
[6.7 3.1 4.7 1.5]: 1 (versicolor)
[6.3 2.3 4.4 1.3]: 1 (versicolor)
[5.6 3. 4.1 1.3]: 1 (versicolor)
[5.5 2.5 4. 1.3]: 1 (versicolor)
[5.5 2.6 4.4 1.2]: 1 (versicolor)
[6.1 3. 4.6 1.4]: 1 (versicolor)
[5.8 2.6 4. 1.2]: 1 (versicolor)
[5. 2.3 3.3 1. ]: 1 (versicolor)
[5.6 2.7 4.2 1.3]: 1 (versicolor)
[5.7 3. 4.2 1.2]: 1 (versicolor)
[5.7 2.9 4.2 1.3]: 1 (versicolor)
[6.2 2.9 4.3 1.3]: 1 (versicolor)
[5.1 2.5 3. 1.1]: 1 (versicolor)
[5.7 2.8 4.1 1.3]: 1 (versicolor)
[6.3 3.3 6. 2.5]: 2 (virginica)
[5.8 2.7 5.1 1.9]: 2 (virginica)
[7.1 3. 5.9 2.1]: 2 (virginica)
[6.3 2.9 5.6 1.8]: 2 (virginica)
[6.5 3. 5.8 2.2]: 2 (virginica)
[7.6 3. 6.6 2.1]: 2 (virginica)
[4.9 2.5 4.5 1.7]: 2 (virginica)
[7.3 2.9 6.3 1.8]: 2 (virginica)
[6.7 2.5 5.8 1.8]: 2 (virginica)
[7.2 3.6 6.1 2.5]: 2 (virginica)
[6.5 3.2 5.1 2. ]: 2 (virginica)
[6.4 2.7 5.3 1.9]: 2 (virginica)
[6.8 3. 5.5 2.1]: 2 (virginica)
[5.7 2.5 5. 2. ]: 2 (virginica)
[5.8 2.8 5.1 2.4]: 2 (virginica)
[6.4 3.2 5.3 2.3]: 2 (virginica)
[6.5 3. 5.5 1.8]: 2 (virginica)
[7.7 3.8 6.7 2.2]: 2 (virginica)
[7.7 2.6 6.9 2.3]: 2 (virginica)
[6. 2.2 5. 1.5]: 2 (virginica)
[6.9 3.2 5.7 2.3]: 2 (virginica)
[5.6 2.8 4.9 2. ]: 2 (virginica)
[7.7 2.8 6.7 2. ]: 2 (virginica)
[6.3 2.7 4.9 1.8]: 2 (virginica)
[6.7 3.3 5.7 2.1]: 2 (virginica)
[7.2 3.2 6. 1.8]: 2 (virginica)
[6.2 2.8 4.8 1.8]: 2 (virginica)
[6.1 3. 4.9 1.8]: 2 (virginica)
[6.4 2.8 5.6 2.1]: 2 (virginica)
[7.2 3. 5.8 1.6]: 2 (virginica)
[7.4 2.8 6.1 1.9]: 2 (virginica)
[7.9 3.8 6.4 2. ]: 2 (virginica)
[6.4 2.8 5.6 2.2]: 2 (virginica)
[6.3 2.8 5.1 1.5]: 2 (virginica)
[6.1 2.6 5.6 1.4]: 2 (virginica)
[7.7 3. 6.1 2.3]: 2 (virginica)
[6.3 3.4 5.6 2.4]: 2 (virginica)
[6.4 3.1 5.5 1.8]: 2 (virginica)
[6. 3. 4.8 1.8]: 2 (virginica)
[6.9 3.1 5.4 2.1]: 2 (virginica)
[6.7 3.1 5.6 2.4]: 2 (virginica)
[6.9 3.1 5.1 2.3]: 2 (virginica)
[5.8 2.7 5.1 1.9]: 2 (virginica)
[6.8 3.2 5.9 2.3]: 2 (virginica)
[6.7 3.3 5.7 2.5]: 2 (virginica)
[6.7 3. 5.2 2.3]: 2 (virginica)
[6.3 2.5 5. 1.9]: 2 (virginica)
[6.5 3. 5.2 2. ]: 2 (virginica)
[6.2 3.4 5.4 2.3]: 2 (virginica)
[5.9 3. 5.1 1.8]: 2 (virginica)
from sklearn.model_selection import train_test_split
# Random state ensures reproducibility
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.33, random_state=42
)
print(f"Training data size: {len(X_train)}")
print(f"Test data size: {len(X_test)}")
Training data size: 100
Test data size: 50
from sklearn.metrics import ConfusionMatrixDisplay
import matplotlib.pyplot as plot
def display_confusion_matrix(classifier, title):
disp = ConfusionMatrixDisplay.from_estimator(
classifier,
X_test,
Y_test,
display_labels=data.target_names,
cmap=plot.cm.Blues,
normalize="true",
)
disp.ax_.set_title(title)
plot.show()
## Classifier
from sklearn import svm, metrics
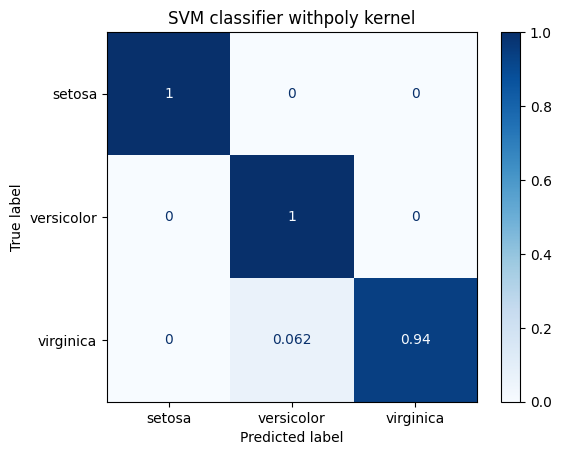
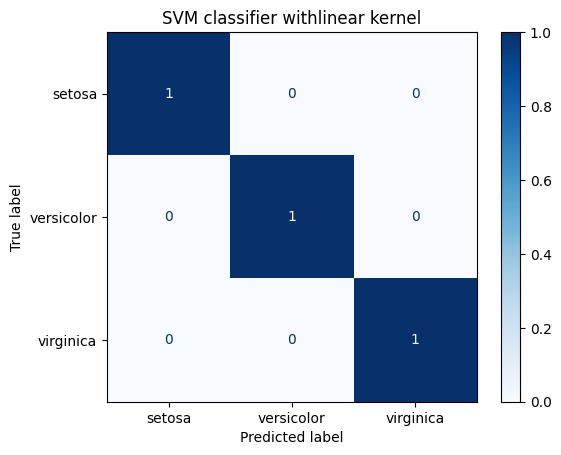
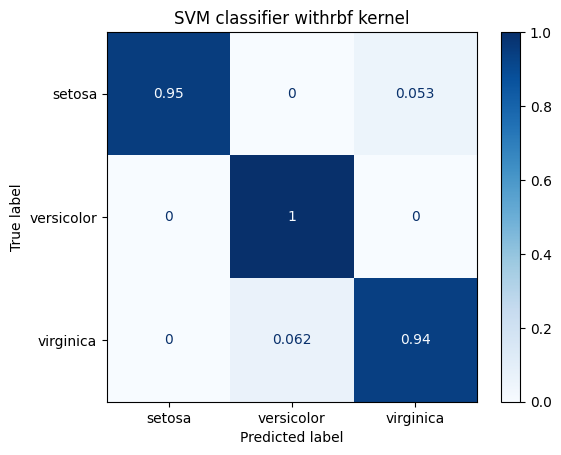
for kernel in ["linear", "rbf", "poly"]:
print("")
classifier = svm.SVC(kernel=kernel, gamma=10)
classifier.fit(X_train, Y_train)
predicted_values = classifier.predict(X_test.reshape((X_test.shape[0], -1)))
# classification report
title = "SVM classifier with" + kernel + " kernel"
print(metrics.classification_report(Y_test, predicted_values))
display_confusion_matrix(classifier, title)
precision recall f1-score support
0 1.00 1.00 1.00 19
1 1.00 1.00 1.00 15
2 1.00 1.00 1.00 16
accuracy 1.00 50
macro avg 1.00 1.00 1.00 50
weighted avg 1.00 1.00 1.00 50
precision recall f1-score support
0 1.00 0.95 0.97 19
1 0.94 1.00 0.97 15
2 0.94 0.94 0.94 16
accuracy 0.96 50
macro avg 0.96 0.96 0.96 50
weighted avg 0.96 0.96 0.96 50
precision recall f1-score support
0 1.00 1.00 1.00 19
1 0.94 1.00 0.97 15
2 1.00 0.94 0.97 16
accuracy 0.98 50
macro avg 0.98 0.98 0.98 50
weighted avg 0.98 0.98 0.98 50